Dortmund, 6th August 2021
The term neuromuscular diseases (NMD), commonly known as muscular atrophy, encompasses all diseases that affect the interaction of nerves and muscles. They can be of a genetic or idiopathic (without an identifiable cause) nature and are very rare and incurable. These diseases, which include polyneuropathies, spinal muscular atrophies, and Duchenne muscular dystrophy, are characterised by a progressive weakness of the skeletal muscles and, frequently, of the cardiac or respiratory muscles.
The diagnosis often comes late, which delays or even prevents treatment. This limits the quality of life of those affected and increases their mortality. A more precise understanding of the molecular causes of these NMDs is needed in order to improve the treatment options, which are currently limited. Gene and protein signatures can revolutionise the diagnosis of NMD patients and improve genetic counselling. Several research groups (Proteomics and Translational Analytics) at ISAS are therefore collaborating intensively on NMD research.
Prediction of genetic defects & new classification
The researchers are making use of muscle biopsies from patients to generate new proteomic and morphological data by applying omics technologies. In the first step, they quantify the causative (i.e. pathogenic) proteins, and in the second step, they compare the results of the measurement with the results of the DNA analysis. The data from ISAS then becomes part of an algorithm. "This algorithm has made it possible, for the first time, to identify patterns of gene and protein co-regulation in NMDs, allowing the pathogenic genetic defect to be narrowed down much more precisely, if not predicted exactly", explains Dr Andreas Hentschel, a member of the Translational Analytics research group. In conjunction with histological-biochemical characterisation by means of biospectroscopy, this allows a new classification of hereditary and acquired NMD disorders. This facilitates the tracing of the underlying pathomechanism, the causal chain of bodily processes leading to disease. This is particularly important for the testing of new therapies.

Dr. Andreas Hentschel is investigating patterns of gene and protein co-regulation in order to narrow down the genetic defect in neuromuscular diseases more precisely than was previously possible.
What are Omics Technologies?
The term omics is used in research to describe molecular biological methods, for example genomics, lipidomics, metabolomics or proteomics, with which biomolecules from tissue samples or other biological samples can be studied on a global level. Omics technologies are an important starting point in personalised medicine (precision medicine), as they produce large amounts of data that provide information about disease processes and potential therapeutic approaches.
Established procedure & increased efficiency
In 2020, the researchers processed approximately 300 samples from patients with different genetic neuromuscular diseases and subsequently examined 100 of them using global proteomics. "We achieved our annual goal of completing the analyses using global proteomics. We also mastered the challenge of quickly tailoring the purification of the samples, as they originate from many different hospitals," summarises Hentschel. The mass spectrometric measurements were performed in 'data independent acquisition' (DIA) mode. The scientists evaluated the results using the spectra library they had previously created. In this way, they were able to increase the efficiency of the analysis by approx. 10 to 40 percent compared to the previously common procedure, data dependent acquisition (DDA). In addition, scientists began to further their research into idiopathic NMD disorders. By the end of the year, the inventory already included 140 samples.
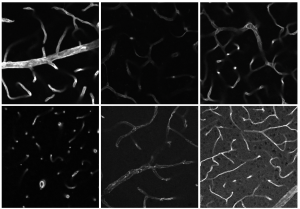
Using CARS microscopy, ISAS researchers have examined more than 80 patient samples (160 muscle biopsy sections in total) from various disease entities since the project started in 2019. In the process, they made overview measurements and detailed measurements. The researchers recorded the muscle fibre calibres of 73 patient samples, evaluated the spectra of 40 samples and used computer-assisted methods for 34 samples. These include the description of intensity ratios between different wavelengths and non-linear unmixing according to Heylen et al. With this unmixing, "pure" spectra are unmixed from the raw spectra, which are usually mixtures of spectra from different substances, and the abundances based on these spectra are displayed in the image or recording.
Targeted assays established
The researchers have succeeded in establishing a targeted assay, a standardised reaction procedure for the detection of gene and protein signatures, using LC-MS-MS (liquid chromatography in combination with mass spectrometry), which has a high degree of sensitivity and specificity. At the same time, the researchers developed another method that considerably shortens the measurement time. They have already used both assays on patient samples to detect the regulation of 74 neuromuscularly relevant proteins.
They also worked on an additional 110 proteins out of the total of 380 known proteins of neuromuscular relevance in 2020. The scientists have already completed the pre-selection and verification of the proteotypic peptides of these proteins and could start the synthesis of the isotopically labelled standard peptides in 2020.
(Pauline Jürgens)
The project entitled, 'Gene and Protein Signatures as GPS for Patients with Neuromuscular Diseases' was funded by the State of North Rhine-Westphalia using resources from the 2014-2020 European Regional Development Fund (ERDF), "Investment for Growth and Jobs" (funding number ERDF-0801301).